Please click term for full definition
Autonomously replicating sequence (ARS):
Sequence that confers the abilty on
plasmids to replicate autonomously of the host
chromosomes, by acting as an origin of replication
(usually used to refer to sequences from yeast).
Sequences from yeast chromosomes that function as
ARS elements may also function as
origins in their original chromosomal
location.
ARS consensus sequence (ACS):
Consensus sequence found in S. cerevisiae origins
of DNA replication.
Checkpoint mechanism
Regulatory mechanism that arrests cell cycle when
previous stages have not been completed or when a
chromosome or DNA defect is present
Clamp loader
Protein such as replication factor C that can load
a clamp such as PCNA onto DNA
Elongation
Second stage of DNA synthesis, following
initiation, where bulk DNA synthesis takes place on
the leading and lagging strands
Fork regression
Occurs when a stalled replication fork reverses
allowing the nascent strands to anneal.
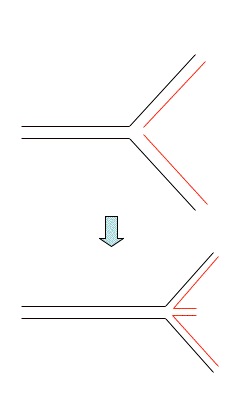

Fragile site
Chromosomal site in mammalian chromosomes that is
prone to breakage.
Initiation
First stage of DNA synthesis where the DNA double
helix is unwound and an initial priming event by
DNA polymerase alpha occurs on the leading strand.
Priming event on lagging strand establishes a
replication fork
Lagging strand
The strand which is synthesised discontinuously
during elongation.
Leading strand
The strand which is synthesised continuously during
elongation.
Licensing
Association of Mcm2-7 proteins with DNA at ORC to
form a pre-replicative complex (pre-RC) . Step
requires Cdc6 and Cdt1. Synonomous with pre-RC
formation
Mcm paradox
Usually refers to the high ratio of Mcm2-7 to ORC
complexes (>10:1) at origins.
Has also been used to refer to the situation where MCMs do not apparently localise with replication sites.
Has also been used to refer to the situation where MCMs do not apparently localise with replication sites.
ori
Abbreviation for origin of DNA replication
Origin efficiency
Probability that activation of DNA replication from
an origin will occur in a single S phase. Origin
efficiency can be high (>80%) in S. cerevisiae,
but appears to be lower in S. pombe and mammalian
cells.
Origin suppression
Origin suppression refers to the inhibition of
initiation at an origin by initiation at a nearby
origin. Can occur passively, whereby passage of a
replication fork through the unfired origin
prevents initiation.
Okazaki fragment
The unit in which DNA is synthesized
discontinuously on the lagging strand. Okazaki
fragments are subsequently ligated together.
Polymerase switching
Process whereby DNA polymerase delta or epsilon
takes over from DNA polymerase alpha. Occurs once
on leading strand on initiation and is required for
generation of each Okazaki fragment
Pre-initiation complex (pre-IC)
Originally used to refer to complex of Sc Cdc45
with pre-RCs before initiation. Now generally used
to refer to more extensive complex of proteins at
origin after pre-RC formation but before initiation
of DNA synthesis.
Pre-replicative complex (pre-RC)
Originally defined as an extended in vivo footprint
on S. cerevisiae origins seen in G1 phase. Now
generally used to refer to origins where Mcm2-7
(together with Cdc6 and possibly Cdt1) is chromatin
associated.
Priming
Synthesis of an RNA primer to allow DNA synthesis
by DNA polymerase alpha. Occurs once at the origin
on the leading strand and at the start of each
Okazaki fragment on the lagging strand.
Processivity
Ability of a DNA polymerase to synthesize
consecutive nucleotides without dissociating from
the template
Proofreading
Ability of DNA polymerases to remove incorrectly
incorporated nucleotides (i.e. where the A-T, C-G
base pairing rules are not observed), via transient
activation of a 3’-5’ exonuclease activity
Replication focus
Cytologically defined site in nucleus where DNA
synthesis is occuring
Replication fork
A site of active DNA synthesis
Replication fork barrier
Protein complex that prevents passage of
replication fork in one direction
Replication origin
DNA sequence where initiation of replication
occurs. In S. cerevisiae origins are sequence
specific, but this appears not to be the case in
most eukaryotes
Replication origin timing
Refers to the fact that replication origins can
show a defined temporal pattern of activation so
that individual origins can fire early or late in S
phase.
Replicon hypothesis
Concept of DNA replication proposed by Brenner,
Jacob and Cuzin. DNA replication was suggested to
initiate at a specific origin via interaction with
an origin binding protein. The unit of replication
replicated from this site is the replicon.
S phase
Phase of cell cycle when DNA synthesis
occurs